A research paper entitled "Integrative analysis of reference epigenomes in 20 rice varieties" published online in Nature Communications on May 27, 2020. Prof. Xingwang Li and Prof. Guoliang Li from National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University (HZAU) are co-corresponding authors, while Dr. Lun Zhao, a postdoctoral researcher of HZAU, and Liang Xie and Qing Zhang, PhD students of this team, are co-first authors.

A previous research results published in Nature Communications by Prof. Xingwang Li and Guoliang Li’s team in 2019 revealed the rice high-resolution three-dimensional genome map (https://rdcu.be/b4rup). This study depicted comprehensive epigenome maps of 20 representative rice varieties and annotated 82% of the functional DNA elements in the rice genome. As parts of the riceENCODE project, these two researches focus on the annotation of functional DNA elements, the spatial genome organization, and their functions on transcriptional regulation and import agronomic traits in rice (from one-dimensional epigenome to 3D genome architecture).
First, they developed a fast and efficient plant ChIP-Seq method (eChIP). The amount of ChIP DNA obtained by eChIP is dozens of times that by the regular ChIP method. eChIP works well for modified histones and transcription factors in both monocotyledon and dicotyledon plants such as rice, Arabidopsis, maize, and Brassica napus.
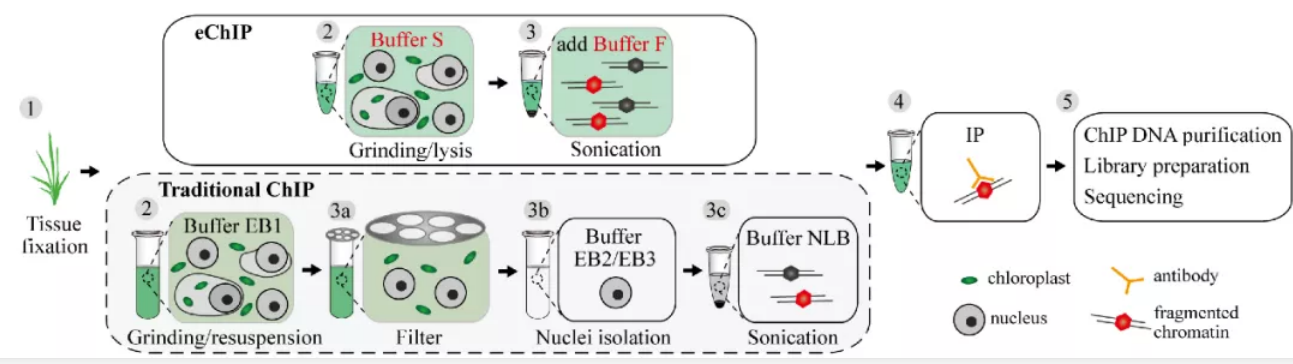
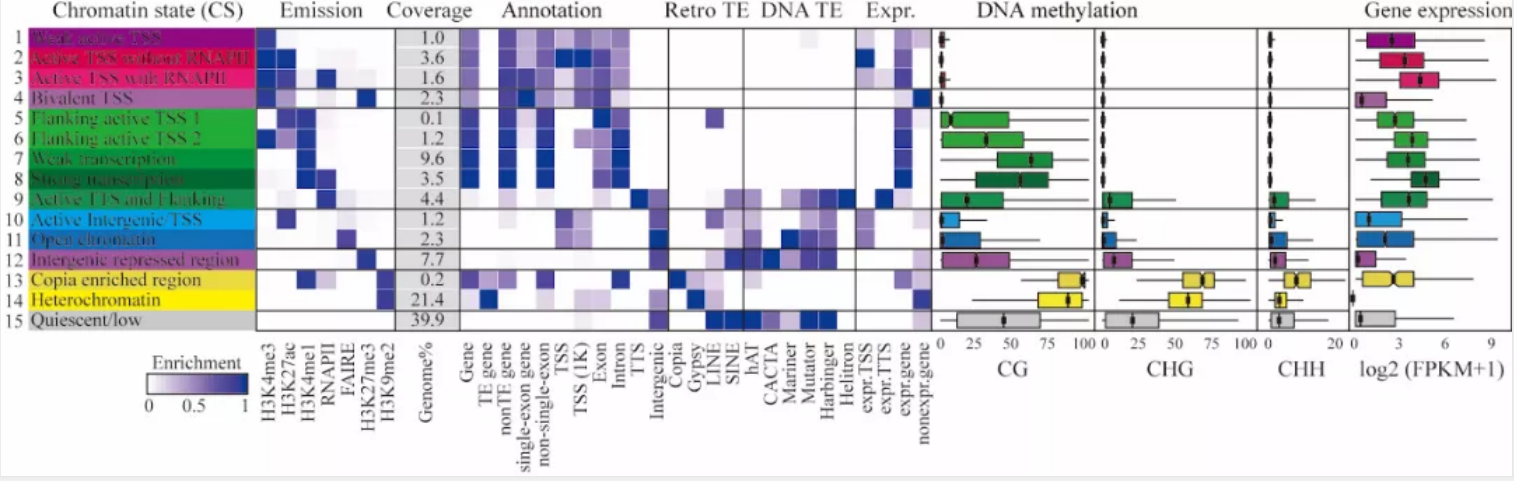
Then, They generated more than 500 datasets across various tissues in 20 representative rice varieties, including 58 gene expression datasets (RNA-Seq) and 32 genome-wide DNA methylation datasets (BS-Seq), 354 histone modification datasets (eChIP-Seq), and 58 open chromatin datasets (FAIRE-Seq). By integrating these data, 15 chromatin states were defined in the rice genome. In addition, a new bivalent chromatin state (H3K9me2/H3K4me1) was identified by ChIP-reChIP. The new bivalent state showed the highest enrichment of the retrotransposon Copia, which frequently located within gene body regions, especially in introns, and the expression of related genes is low,but its potential biological function needs further study.
Furthermore, the researchers refined the genomic region of active promoters: open chromatin region and H3K4me3 modified region, which is approximately 500 bp upstream to 1 kb downstream of the transcription start site. Also, they found the enhancer-like promoters, which not only regulate the expression of proximal genes but also play enhancer-like role in distal genes through chromatin interactions.
Finally, the author conducted a comparative analysis of the epigenome maps among the 20 rice varieties, and found that heterochromatic regions are more prone to have apparent modification variation. At the same time, the study notes important areas of apparent differences between Indica and Japonica rice. These results provide a unique perspective and important datasets for studying the differentiation and environmental adaptability of rice varieties.
In summary, this study provides a comprehensive overview of rice epigenome maps, promoter chromatin characteristics, enhancer-like promoters and epigenetic dynamics in different tissues, and the effects of genetic variation on rice epigenome maps. These datasets also serve as a valuable resources for the studies of rice functional genomics and breeding.
This research was financed by funding from the National Key Research and Development Program, the National Natural Science Foundation of China, Huazhong Agricultural University and the National Key Laboratory of Crop Genetic Improvement.
The research article: https://www.nature.com/articles/s41467-020-16457-5